|
Research Articles
Phenotypic and genotypic characterization of carbapenemsresistant clinical gram-negative bacteria from Sudan
1 Lecturer, Department of Microbiology, Medicine Program, Batterjee Medical College, Jeddah, Saudi Arabia
2 Associate Professor, Department of Microbiology, Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia
3 Lecturer, Laboratory Medicine Department, Faculty of Applied Medical Sciences, Umm Al-Qura University, Saudi Arabia
4 Lecturer, Department of Microbiology, Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia
5 Assistant Professor, Department of Microbiology, Faculty of Medical Laboratory Science, Khartoum University, Sudan
Address correspondence to:
Mohamed Elmutasim Abdelhamid Mohamed
P.O. Box 623, Jeddah 21442,
Saudi Arabia
Message to Corresponding Author
Article ID: 100011M08MM2019
Access full text article on other devices

Access PDF of article on other devices

How to cite this article
Mohamed MEA, Ahmed AO, Kidir ESB, Sirag B, Ali MA. Phenotypic and genotypic characterization of carbapenems-resistant clinical gram-negative bacteria from Sudan. Edorium J Microbiol 2019;5:100011M08MM2019.ABSTRACT
Aims: This study is conducted to characterize the phenotypes and genotypes associated with carbapenems-resistant in gram-negative bacilli isolated from Sudanese patients.
Methods: Three hundred and seventy clinical Sudanese isolates of Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa were screened for meropenem (MEM) resistance by Kirby–Bauer disk diffusion method and Modified Hodge test (MHT). The MEM-resistant phenotype was also confirmed by automated Vitek2® system. Dual-tubed multiplex polymerase chain reaction (PCR) for separate groups of carbapenemase encoding genes (blaNDM-1, blaOXA-181/48, blaVIM, blaKPC, blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58) was examined for MEM-resistant strains.
Results: Screening of the 367 clinical isolates using MEM disk diffusion method resulted in 36 (9.8%) resistant strains. However the most frequently recognized gene was blaOXA-51 27.8% (10/36), pursued by blaNDM-1 and blaOXA-181/48 16.7% (6/36), while blaKPC and blaOXA-23 were less identified 2.7% (1/36). These genes were mostly detected in P. aeruginosa 17 (52%), followed by K. pneumoniae 11 (33%), E. coli 3 (9%), and A. baumannii 2 (6%). The overall rate of concordance between phenotypic and genotypic testing was 82.4% for the detection of carbapenemases by MHT and 70.6% by Vitek2.
Conclusion: In response to the high rate of carbapenemases and carbapenem-resistance encoding genes among Enterobacteriaceae in Sudan, nation-wide antibiotics resistance control and monitoring programs are immediately needed. Based on the finding of this study, Tigecycline can still be used as non-β lactam antibiotic for the treatment of infections caused by carbapenem-resistance gram-negative bacteria.
Keywords: Carbapenems, Carbapenem resistance, Enterobacteriaceae, Gram-negative bacteria
INTRODUCTION
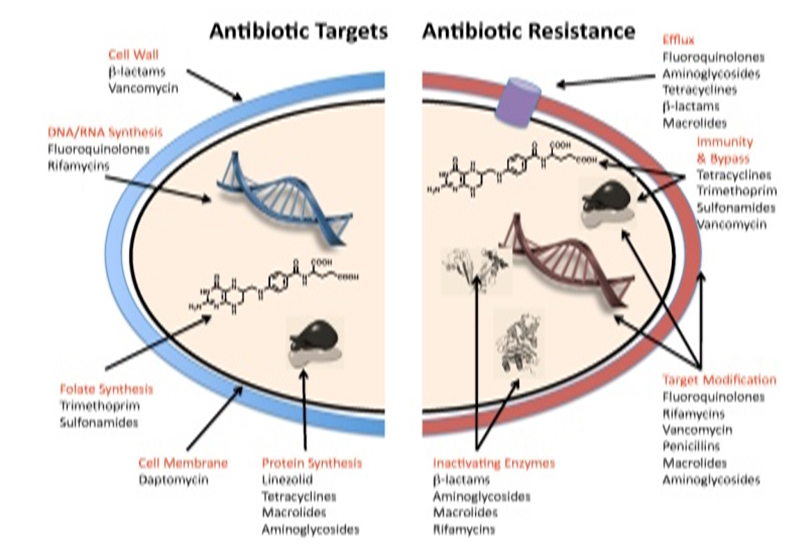
Fermentative and non-fermentative gram-negative bacilli as E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa are common causes of hospital- and community-acquired infections that include cystitis, wound sepsis, pneumonia, peritonitis, bacteremia, and meningitis. Resistance to broad-spectrum antimicrobials, such as the extended-spectrum cephalosporins, is a well-recognized problem among Enterobacteriaceae [1]. Until recently carbapenems have served as an important antimicrobial class for the treatment of these organisms and, resistance to carbapenems has been uncommon among Enterobacteriaceae. However, therapeutic failures are observed due to resistance to carbapenem drugs. Resistance to carbapenems is due to several mechanisms like (i) production of carbapenemases, (ii) reduced expression of outer membrane protein, (iii) multidrug efflux pump, and (iv) altered expression of penicillin-binding protein (Figure 1) [2].
The spread of resistance to carbapenems among Enterobacteriaceae has turned into significant public health issue. In reality, the hospital and community environment accounted for epidemics or endemic states of patients infected or colonized by Enterobacteriaceae that are not susceptible to carbapenems. Carbapenemresistant Enterobacteriaceae (CRE) is resistant to most β-lactams, non-β-lactam antibiotics, which make them resistant to most antibiotics [3],[4],[5]. Since obtained carbapenemases are encoded by transferable genes found in mobile components such as plasmids and transposes, which can disperse between distinct strains and species, interceding carbapenemase resistance is the main treatment in CRE. The incident of either class B metallo-β-lactamases (MBL: IMP, VIM, NDM) or classes A (KPC) and D (OXA-type) serine carbapenemases has been accounted in Europe and around the world, [3],[4],[5],[6],[7],[8]. Also, the concurrence of carbapenem resistance and extended spectrum β-lactamase (ESBL) production has been frequently reported. In addition, the movement of colonized patients, primarily elderly, between hospital wards and long-term care facilities can help spread CRE [3],[8].
A high mortality rate is being reported due to infections caused by CRB because of limited treatment options available. So prevention of CRE transmission and CRE infections have become important public health objectives. This study has been conducted in order to detect phenotypic and genotypic characterization pattern of carbapenems-resistant gram-negative bacteria isolated from Sudan using Vitek2 compact, dual-tubed multiplex PCR and full genome sequencing. To our knowledge this is the first study of its type in Sudan to address the importance of genotypes of carbapenemase-producing bacteria.
MATERIALS AND METHODS
Study area
Clinical isolates of gram-negative bacilli (GNB) were collected from Khartoum the capital city of Sudan from two microbiology laboratories: Microbiology Laboratory of Soba University Hospital and Khartoum Teaching Hospital. Isolates were collected according to the appropriate regulations during study period. The conventional microbiology work was carried out at the Microbiology Laboratory of Batterjee Medical College, Jeddah, Saudi Arabia while Vitek2 testing, PCR and sequencing were performed at Microbiology Research Laboratory, Umm Al Qura University, Makkah, Saudi Arabia.
Study design
This is a cross-sectional descriptive and laboratory based study.
Study population (bacterial isolates)
This study involved 370 of clinical isolates of E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa. All isolates were collected form Khartoum hospitals as described above. Bacterial isolates were identified using conventional microbiological and PCR-based techniques.
All these isolates have been screened for resistance to MEM by Kirby–Bauer disk diffusion method according to The Clinical & Laboratory Standards Institute (CLSI) guidelines [9].
Modified Hodge test
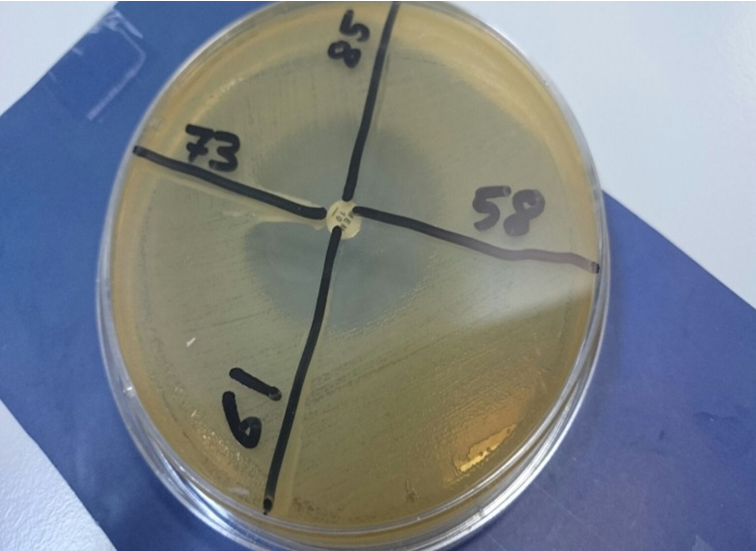
The MEM-resistant strains were subjected to MHT for detection of carbapenemases. An overnight culture suspension of E. coli ATCC 25922 adjusted to 0.5 McFarland standard12 was inoculated using a sterile cotton swab on the surface of a Mueller-Hinton agar (MHA) (Oxoid, UK). After drying, 10 μg MEM disk (Oxoid, UK) was placed at the center of the plate and the test strains were streaked from the edge of the disk to the periphery of the plate in four different directions. The plate was incubated overnight at 37°C. The sample strain showing a cloverleaf shaped inhibition area owing to the manufacturing of carbapenemase was regarded positive (Figure 2). The resistant strains and/or modified Hodge positive were confirmed by using automated Vitek2 system.
Use of Vitek2 for the detection of carbapenems-resistant bacteria
Following manufacturer (BioMérieux) instruction. GN and AST-291 Cards were used for identification of GNB, detection of ESBL production, and carbapenemsresistant bacteria. Antibiotic susceptibility was also tested for Ampicillin (AMP), Amoxicillin-Clavulanic acid (AMC), Piperacillin-Tazobactam (TZP), Cefalotin (CLO), Cefuroxime-Axetil (CAE), Cefoxitin (FOX), Ceftazidime (CAZ), Ceftriaxone (CRO), Cefepime (FEP), Imipenem (IPM), Meropenem (MEM), Amikacin (AMK), Gentamicin (GEN), Ciprofloxacin (CIP), Tigecycline (TGC), Nitrofurantoin (NIT), Trimethoprim-Sulfamethoxazole (SXT). The CLSI 2012 M100-S23 breakpoint values were used for results interpretation [9].
DNA extraction
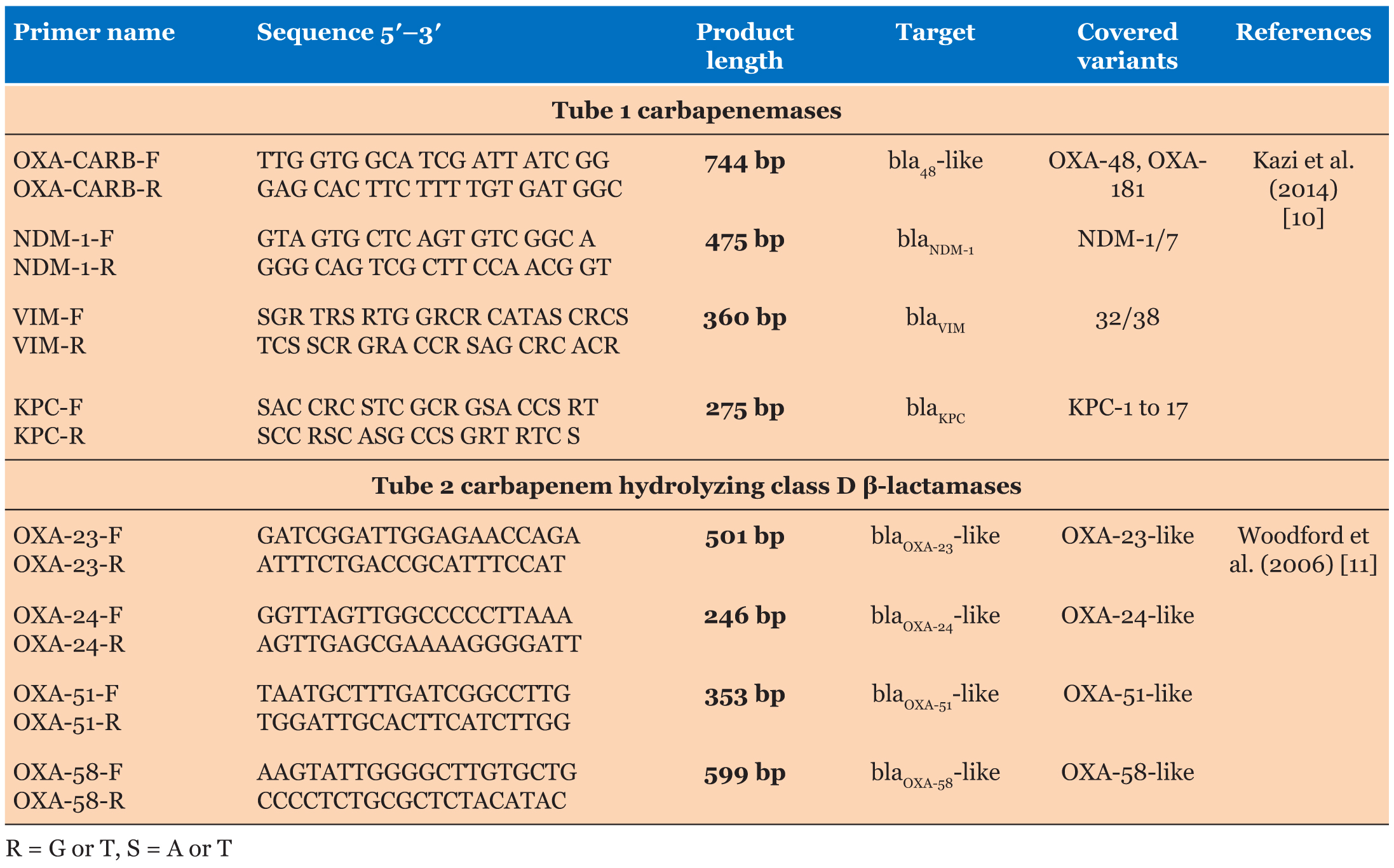
Total DNA was extracted from 36 carbapenemsresistant isolates using simple boiling technique. Two colonies of bacteria were picked from a plate and emulsified into 50 μL of TE buffer. The bacterial suspension was boiled at 100°C for 10 minutes. After boiling, the bacterial lysate was centrifuged at 14,000 rpm for 10 minutes and the supernatant was used as DNA template for PCR amplifications as described by Kazi et al. [10]. The primers used in this study were obtained from Integrated DNA Technologies. Table 1 shows the detailed information of the primer sets used.
Dual-tubed multiplex PCR
Two tubes multiplex PCR was performed to detect the following targets: 1. Tube 1(blaNDM-1, blaOXA-181/48, blaVIM, and blaKPC). 2. Tube 2 (blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58).
Targets were amplified using PCR with initial denaturation of 94°C for 3 minutes, followed by 34 cycles of 94°C for 30 seconds, 45.3°C for 30 seconds, 72°C for 30 seconds, followed by 34 cycles of 94°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds, with a final extension of 72°C for 6 minutes. Postamplification products were visualized on 1.5% agarose gel electrophoresis.
Ethics statement
Only bacterial isolates recovered and leftover from routine diagnostic laboratory were used in this study without a direct use of clinical specimens. No name data or patients ID were linked with the isolates and therefore patient consents were not required. The study was ethically approved by committee of Faculty of Medical Laboratory Sciences, University of Khartoum, Sudan.
RESULTS
Phenotypic analysis
Of 367 clinical isolates screened by MEM disk diffusion method only 36 (9.8%) were found to be resistant to MEM (MIC ≥2 mg/mL). Among these 27% (10/36), 16% (6/36), 38.8% (14/36), and 2% (1/36) were E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii, respectively. However, when these 36 CRE isolates were analyzed with MEM-MHT; only 26 (72%) isolates were found positive with MEM-MHT as example in Figure 2.
In antimicrobial susceptibility testing using VITEK AST-N291 cards 50% (18/36) showed intermediate or resistance designations to imipenem, MEM or both. Phenotype detected by VITEK when all 36 CRE isolates were considered; showed 100% were resistant to Ampicillin, Amoxicillin/Clavuanic acid, Ceftazidime, Ceftriaxone, and Cefepime. In these 36 isolates 88% were resistant to Cefoxitin, 85% were resistant to Nitrofurantoin and 70% were resistant to Ciprofloxacin. Almost all of the isolates (97%) were found to be sensitive to Tigecycline except two P. aeruginosa isolates. Twentyfive of the 36 isolates (70%) were multidrug resistant which was defined as being resistant to three or more functional classes of drugs simultaneously by Magiorakos et al. [12]. Most of ESBLs (80%) were found among E. coli.
Genotypic analysis
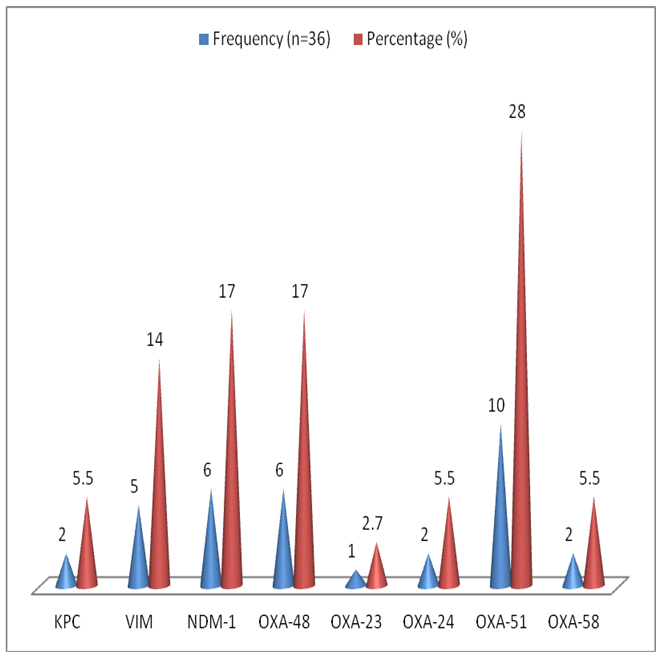
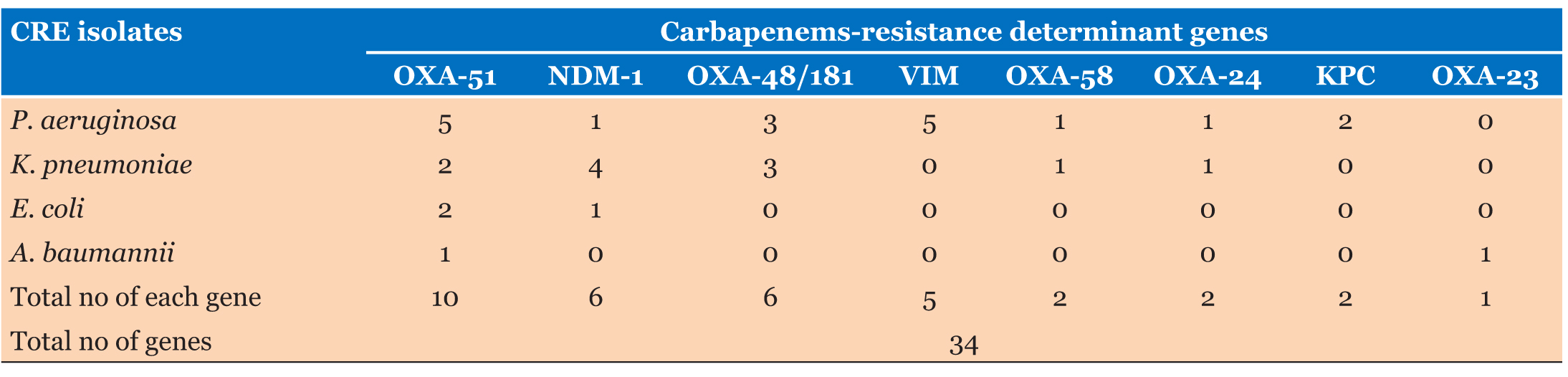
Based on PCR, 18 (50%) of 36 multidrug resistance (MDR) gram-negative bacteria isolates were positive for one or more of the carbapenem-resistance determine genes. OXA-51 β-lactamase genes were the most frequent genes, which were detected in 10 of 36 (MDR) bacterial isolates, followed by OXA-48 and New Delhi metallo-β-lactamase (NDM-1) in six isolates, each of 36 (MDR) bacterial isolates and OXA-23 was the least found in six isolates of 36 (MDR) bacterial isolates. More details are shown in Figure 3.
Based on PCR detected genes among 18 MDR clinical isolates, only blaNDM-1 and blaOXA-51 were found as single carbapenemase resistant genes in four different MDR clinical isolates, while multiple carbapenemase-resistant genes were detected in all other 14 MDR clinical isolates. Genotypic distribution of carbapenemases among K. pneumoniae, E. coli, P. aeruginosa, and A. baumannii is presented in Table 2.
Among these CRE bacterial isolates, the most frequently detected gene was OXA-51 28% (10/36), followed by NDM-1 and OXA-48/181 16.7% (6/36), while KPC and OXA-23 were less detected 2.7% (1/36).
In general, carbapenems-resistance determinant genes were found in 50% (18/34) of P. aeruginosa, followed by 32% (11/34) of K. pneumoniae. Known carbapenems resistance genes were found in only 9% (3/34) of E. coli and 6% (2/34) of A. baumannii. The most frequent carbapenems resistance genes among P. aeruginosa were VIM and OXA-51, 15% (5/34) in each. While most frequently detected gene among K. pneumoniae was NDM-1, which was seen in 11% (4/36) of the isolates.
The overall rate of concordance between phenotypic and genotypic testing was 82.4% (14/17) for the detection of carbapenemases by MHT and 70.6% (12/17) by Vitek2 AST-N291 cards.
DISCUSSION
Extended spectrum β-lactamase producers can be treated with combinations of third generation cephalosporin’s and β-lactamase inhibitors, but ampC producers require carbapenem use, particularly in systemic infections. Extended spectrum β-lactamases in Enterobacteriaceae are reported to coexist with resistance to other antimicrobial classes and as such these organisms become multidrug resistant hence limiting treatment options for infections.
In Sudan the frequency of ESBL producing isolates varied between hospitals (18.2–45.1%), although Khartoum Teaching Hospital had a large incidence of 45.1% [13]. Extended spectrum β-lactamase producers have been reported to as well harbor carbapenemresistance determining genes in 50% of cases in South Africa [14]. Among the isolates from Sudan, this data is limited. Moreover, among the MDR strains, the data is even more limited.
This study was conducted in order to detect phenotypic and genotypic characterization pattern of carbapenemsresistant gram-negative bacteria isolated from Sudan. To our knowledge this is the first study of its type in Sudan to address the importance of genotypes of carbapenemasesproducing bacteria.
This study has revealed a prevalence of the carbapenamase phenotype of 9.8% using the MEM disk diffusion method and/or MHT and genotype of 5% among MDR K. pneumoniae, E. coli, P. aeruginosa, and A. baumannii.
The phenotypic incidence of MHT screening was very large compared to 2.8% in Morocco [15]. The distinction in these discoveries could be on the grounds that the information from Morocco originated from epidemiological investigations of CRE episodes in a emergency clinic setting. This study prevalence is also much higher than that obtained in studies from China and Germany [16],[17], as well as in a surveillance study in Spain which reported carbapenemase encoding gene prevalence of 0.04% [18]. These differences can be attributed to the unrestricted use of antibiotics in Sudan, which plays an important role in increasing carbapenem resistance. These study findings are lower to those observed in Nigeria, Uganda, Tanzania, and in neighboring Egypt where studies reported a prevalence of 33.5, 28.9, 35, and 62.7%, respectively [19],[20],[21],[22].
Modified Hodge test is recommended for the carbapenemase phenotype identification test by CLSI [9]. Studies have shown that MHT is known to be reliable for the identification of KPCs and enzymes similar to OXA- 48 but poorly performed for MBLs (NDMs, VIMs, and IMPs) [23],[24]. However, in this research, we discovered that all beneficial MBL (NDM1 and VIM) bacteria were positive for MHT. The findings of this study were comparable to those observed in Turkey [25]. In this study, the positive rate of MHT was 26/36, indicating the occurrence of false negative, which might be associated with the type B carbapenemase that can be detected by ethylenediaminetetraacetic acid (EDTA) synergy test. Also, there is limited data on MHT’s performance for CRE other than carbapenem-resistant K. pneumoniae and E. coli. Hammoud et al. showed MHT gave false-positive results in Enterobacter cloacae [26]. In this regard, this study analyzed performance of MHT on many different fermentative and non-fermentative gram-negative bacilli CRE species. In this study MHT performed best results for blaNDM-1, blaKPC, blaVIM, blaOXA-48/181 and isolates carrying multiple genes for K. pneumoniae and P. aeruginosa. However, MHT performed poorly for A. baumannii and E. coli.
The present study revealed that the overall detection rate by MHT was 72%. These findings are however comparable to those observed in Egypt, and another Indian study, in which by using MHT test, 80% of carbapenem-resistant K. pneumoniae isolates can be detected [23],[27]. Moreover, Amjad et al. reported that MHT detection rate of 69% [28]. In contrast with these findings, an Ugandan study showed a relatively low MHT carbapenamase detection rate of 10.2% [15]. Similarly, a Moroccan study showed carbapenemase production by MHT to be 2.8% [15]. This may be explained by that false negative results that associated with the test method instead of the type of carbapenemases [29]. Wang et al. have reported a false positive rate of 3.3% in MHT due to the low ertapenem hydrolysis capacity of ESBL, especially CTX-M [30]. Therefore, a combination examination of carbapenemase phenotype by MHT and EDTA synergy test can improve the sensitivity and specificity of carbapenemase gene detection.
In this study, most of the tested isolates were resistant to first line drugs. Most of the CRE are multidrug resistant, but may remain susceptible to Tigecycline. A recent study indicated that Tigecycline has a similar efficacy to other antibiotics in treating CRE infections. Combination therapy and high-dose regimens may be superior to monotherapy and standard-dose regimens, respectively [31].
blaOXA-51 carbapenemase gene was detected and codetected in a surprising number of strains as the most predominant gene among the 36 CRE isolates at a rate of 27%. As these genes were thought to reside exclusively in Acinetobacter species only. The blaOXA- 51-like genes were regarded to be solely chromosomally encoded, intrinsic oxacillinase genes of A. baumannii and are used for the identification of species and strain typing by many researchers [32],[33],[34]. A number of latest reports, however, show that the blaOXA-5-like genes were mobilized and spread by conjugative plasmids to other Acinetobacter species [35],[36]. A study for isolates from Sierra Leone bolsters this contention and corroborates in a meeting abstract that describes the presence of blaOXA- 51-like and blaOXA-58-like genes in K. pneumoniae and E. coli isolates, respectively [37]. However, in this study we detected blaOXA-51 as the most determining genes among P. aeruginosa, also it had been found among A. baumannii, K. pneumoniae, and E. coli isolates.
The most common CRE species isolated in the United States and European countries are K. pneumoniae followed by P. aeruginosa and E. coli [23],[38]. In accordance to this study, the most common isolated species was P. aeruginosa, among which the predominant genes were blaOXA-51 and blaVIM. However, the second most common resistant gene was NDM-1 in K. pneumoniae. This data similar to the findings of a study done in Korea where VIM was reported as the most predominate CRE in class B MBL between gram-negative clinical isolates. It also agrees with the results that report VIM as the most common MBL-resistant gene globally [39].
Plasmid carrying carbapenemase gene like NDM is diverse and can harbor a high number of resistance genes associated with other carbapenemase genes like oxacillinase 48 types, VIM types, ESBL genes, etc. as the source of multidrug resistance in one bacteria [40]. This study detected a low prevalence rate of the blaNDM-1 gene 1.6% of 367 isolates. This finding is also similar to what observed in Tanzania, where it was seen in only 3.1% of 227 isolates [41]. Furthermore, the blaNDM-1 gene coexisted in the same isolate with at least one other carbapenamase genes in three of five isolates among which it was detected. This probably explaining why these latter isolates were multidrug resistant.
Of 36 CRE isolates 34% had multiple genes coding for carbapenem resistance. This data is comparable to study done in Tanzania that found similar results at 32% of the isolates [41]. This is probably due to study of eight most common genes in our research. Sudan has a geographical importance for isolation of carbapenemase-producing bacteria. Manenzhe et al. (2014) revealed about 83 studies in Africa showing that the incidence in hospital environments of carbapenemase producer isolates ranged from 2.3 to 67.7% in North Africa and from 9 to 60% in sub-Saharan Africa [42]. However, molecular studies regarding carbapenemase resistance are limited. The overall rate of concordance between phenotypic and genotypic testing was 82.4% (14/17) for the detection of carbapenemases by MHT and 70.6% (12/17) by Vitek2 AST-N291 cards.
In this study, all NDM-1, KPC, VIM, and OXA-48/181 positive isolates were positive with MHT, with 100% sensitivity. Isolates carrying multiple genes were almost all detected by the MHT method, with sensitivity 92%. However, MHT fails to detect OXA-51 when it is found as a single gene. Regarding Vitek2 AST-N291 cards it showed good results in detection of MBL genes or combination of MBLs and carbapenem hydrolyzing class D β-lactamases (CHDLs) genes, with sensitivity 91%. However, it fails to detect 50% of CHDL genes.
These finding are similar to those observed in other automated antibiotic susceptibility studies, which found that automated systems either over or under reporting carbapenem resistance [23],[24]. In the event of atypical strains or hospital outbreaks, however, these phenotypic information generally have restricted resolution needed to understand complicated resistant phenotypes, outbreak clones, colonization dynamics, and badly differentiated organism species identity. This finding encourages more uses of molecular methods for CRE diagnosis.
CONCLUSION
This study has demonstrated a high prevalence of carbapenemases and carbapenem-resistance encoding genes in fermentative and non-fermentative gram-negative bacilli clinical isolated collected from Microbiology Laboratory of Soba University Hospital and Khartoum Teaching Hospital in Sudan. All the eight genes tested (blaNDM-1, blaVIM, blaKPC, blaOXA-48-like, blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58) were detected in the study sample. The most prevalent gene was blaOXA-51 and the least were blaKPC and blaOXA-23. In this study, almost 10% of the isolates tested were phenotypically and genotypically positive for carbapenemase activity, which is alarming situation and require immediate action for adopting routine screening of carbapenem resistance in Enterobacteriaceae.
REFERENCES
1.
Schwaber JM, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: A potential threat. JAMA 2008;300(24):2911–3. [CrossRef]
[Pubmed]

2.
Wright GD. Bacterial resistance to antibiotics: Enzymatic degradation and modification. Adv Drug Deliv Rev 2005;57(10):1451–70. [CrossRef]
[Pubmed]

3.
Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol Med 2012;18(5):263–72. [CrossRef]
[Pubmed]

4.
Cantón R, Akóva M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 2012;18(5):413–31. [CrossRef]
[Pubmed]

5.
Temkin E, Adler A, Lerner A, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: Biology, epidemiology, and management. Ann NY Acad Sci 2014;1323:22–42. [CrossRef]
[Pubmed]

6.
Miriagou V, Tzelepi E, Gianneli D, Tzouvelekis LS. Escherichia coli with a self–transferable, multiresistant plasmid coding for metallo-beta-lactamase VIM-1. Antimicrob Agents Chemother 2003;47(1):395–7. [CrossRef]
[Pubmed]

7.
Mataseje LF, Boyd DA, Willey BM, et al. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring bla(KPC) from New York City and Toronto. J Antimicrob Chemother 2011;66(6):1273–7. [CrossRef]
[Pubmed]

8.
Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 2015;28(3):565–91. [CrossRef]
[Pubmed]

9.
10.
Kazi M, Drego L, Nikam C, et al. Molecular characterization of carbapenem-resistant Enterobacteriaceae at a tertiary care laboratory in Mumbai. Eur J Clin Microbiol Infect Dis 2015;34(3):467–72. [CrossRef]
[Pubmed]

11.
Woodford N, Ellington MJ, Coelho JM, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006;27(4):351–3. [CrossRef]
[Pubmed]

12.
Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18(3):268–81. [CrossRef]
[Pubmed]

13.
Ibrahim ME, Bilal NE, Magzoub MA, Hamid ME. Prevalence of extended-spectrum ß-lactamases-producing Escherichia coli from Hospitals in Khartoum State, Sudan. Oman Med J 2013;28(2):116–20. [CrossRef]
[Pubmed]

14.
Brink AJ, Coetzee J, Clay CG, et al. Emergence of New Delhi metallo-beta-lactamase (NDM-1) and Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa. J Clin Microbiol 2012;50(2):525–7. [CrossRef]
[Pubmed]

15.
16.
Hu L, Zhong Q, Shang Y, et al. The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol Infect 2014;142(9):1972–7. [CrossRef]
[Pubmed]

17.
Ehrhard I, Karaalp AK, Hackel T, et al. Prevalence of carbapenemase-producing bacteria in hospitals in Saxony, Germany. [Article in German]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2014;57(4):406–3. [CrossRef]
[Pubmed]

18.
Miró E, Agüero J, Larrosa MN, et al. Prevalence and molecular epidemiology of acquired AmpC ß-lactamases and carbapenemases in Enterobacteriaceae isolates from 35 hospitals in Spain. Eur J Clin Microbiol Infect Dis 2013;32(2):253–9. [CrossRef]
[Pubmed]

19.
20.
Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J Multidiscip Healthc 2013;6:303–10. [CrossRef]

21.
Gwimile JJ, Shekalaghe SA, Kapanda GN, Kisanga ER. Antibiotic prescribing practice in management of cough and/or diarrhoea in Moshi Municipality, Northern Tanzania: Cross-sectional descriptive study. Pan Afr Med J 2012;12:103.
[Pubmed]

22.
Amer WH, Khalil HS, Ali Abd EL Wahab MA. Risk factors, phenotypic and genotypic characterization of carbapenem resistant Enterobacteriaceae in Tanta University Hospitals, Egypt. Int J Infect Control 2016;12(2):1–11. [CrossRef]

23.
Pollett S, Miller S, Hindler J, Uslan D, Carvalho M, Humphries RM. Phenotypic and molecular characteristics of carbapenem-resistant Enterobacteriaceae in a health care system in Los Angeles, California, from 2011 to 2013. J Clin Microbiol 2014;52(11):4003–9. [CrossRef]
[Pubmed]

24.
Solanki R, Vanjari L, Subramanian S, B A, E N, Lakshmi V. Comparative evaluation of multiplex PCR and routine laboratory phenotypic methods for detection of carbapenemases among gram negative bacilli. J Clin Diagn Res 2014;8(12):DC23–6. [CrossRef]
[Pubmed]

25.
Baran I, Aksu N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann Clin Microbiol Antimicrob 2016;15(1):20. [CrossRef]
[Pubmed]

26.
Hammoudi D, Ayoub Moubareck C, Aires J, et al. Countrywide spread of OXA-48 carbapenemase in Lebanon: Surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Int J Infect Dis 2014;29:139–44. [CrossRef]
[Pubmed]

27.
Bina M, Pournajaf A, Mirkalantari S, Talebi M, Irajian G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae isolated from the clinical samples by the phenotypic and genotypic methods. Iran J Pathol 2015;10(3):199–205.
[Pubmed]

28.
Amjad A, Mirza Ia, Abbasi S, Farwa U, Malik N, Zia F. Modified Hodge test: A simple and effective test for detection of carbapenemase production. Iran J Microbiol 2011;3(4):189–93.
[Pubmed]

29.
Azimi L, Talebi M, Owlia P, et al. Tracing of false negative results in phenotypic methods for identification of carbapenemase by Real-time PCR. Gene 2016;576(1 Pt 1):166–70. [CrossRef]
[Pubmed]

30.
Wang P, Chen S, Guo Y, et al. Occurrence of false positive results for the detection of carbapenemases in carbapenemase-negative Escherichia coli and Klebsiella pneumoniae isolates. PLoS One 2011;6(10):e26356. [CrossRef]
[Pubmed]

31.
Ni W, Han Y, Liu J, et al. Tigecycline treatment for carbapenem-resistant Enterobacteriaceae infections: A systematic review and meta-analysis. Medicine (Baltimore) 2016;95(11):e3126. [CrossRef]
[Pubmed]

32.
Patel G, Bonomo RA. Status report on carbapenemases: Challenges and prospects. Expert Rev Anti Infect Ther 2011;9(5):555–70. [CrossRef]
[Pubmed]

33.
Héritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother 2005;49(10):4174–9. [CrossRef]
[Pubmed]

34.
Merkier AK, Centrón D. bla(OXA-51)-type beta-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int J Antimicrob Agents 2006;28(2):110–3. [CrossRef]
[Pubmed]

35.
Chen TL, Lee YT, Kuo SC, et al. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother 2010;54(11):4575–81. [CrossRef]
[Pubmed]

36.
Lee YT, Kuo SC, Chiang MC, et al. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob Agents Chemother 2012;56(2):1124–7. [CrossRef]
[Pubmed]

37.
Leski TA, Bangura U, Jimmy DH, et al. Identification of bla
38.
Glasner C, Albiger B, Buist G, et al. Carbapenemase-producing Enterobacteriaceae in Europe: A survey among national experts from 39 countries, February 2013. Euro Surveill 2013;18(28). pii: 20525. [CrossRef]
[Pubmed]

39.
Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: Epidemiology and Prevention. Clin Infect Dis 2011;53(1):60–7. [CrossRef]
[Pubmed]

40.
Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009;53(12):5046–54. [CrossRef]
[Pubmed]

41.
Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary hospital in Mwanza, Tanzania. Biomed Res Int 2014;2014:303104. [CrossRef]
[Pubmed]

42.
Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa: A systematic review. J Antimicrob Chemother 2015;70(1):23–40. [CrossRef]
[Pubmed]

SUPPORTING INFORMATION
Author Contributions
Mohamed Elmutasim Abdelhamid Mohamed - Conception of the work, Design of the work, Acquisition of data, Analysis of data, Drafting the work, Revising the work critically for important intellectual content, Final approval of the version to be published, Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Abdalla Osman Ahmed - Conception of the work, Design of the work, Acquisition of data, Analysis of data, Drafting the work, Revising the work critically for important intellectual content, Final approval of the version to be published, Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
El-Shiekh Babiker Kidir - Analysis of data, Revising the work critically for important intellectual content, Final approval of the version to be published, Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Bashir Sirag - Analysis of data, Revising the work critically for important intellectual content, Final approval of the version to be published, Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Musa Abdalla Ali - Acquisition of data, Revising the work critically for important intellectual content, Final approval of the version to be published, Agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Guaranter of SubmissionThe corresponding author is the guarantor of submission.
Source of SupportNone
Consent StatementWritten informed consent was obtained from the patient for publication of this article.
Data AvailabilityAll relevant data are within the paper and its Supporting Information files.
Conflict of InterestAuthors declare no conflict of interest.
Copyright© 2019 Mohamed Elmutasim Abdelhamid Mohamed et al. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information.