|
|
Original Article
| ||||||
| Molecular characterization of vancomycin resistant Enterococcus faecium clinical isolate in Makkah, Saudi Arabia | ||||||
| Husam Aldeen Hassan Tahir1, Abdalla Ahmed2, KawtharAbdelgaleil Mohammed Salih3, Bashir Sirag4 | ||||||
|
1Student, College of Graduate Studies, Sudan University of Science and Technology, Khanoum State, Sudan 2Associate Professor, Microbiology Department, Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia 3Assistant Professor, Immunology, Sudan University of Science and Technology, Khanoum State, Sudan 4Lecturer, Microbiology Department, Faculty of Medicine, Umm Al-Qura University, Makkah, Saudi Arabia | ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
[Print This Article] [Similar articles in PubMed] [Similar articles in Google Scholar] |
| How to cite this article |
| Tahir HAH, Ahmed A, Salih KAM, Sirag B. Molecular characterization of vancomycin resistant Enterococcus faecium clinical isolate in Makkah, Saudi Arabia. Edorium J Microbiol 2018;4:100009M08HT2018. |
|
ABSTRACT
| ||||||
|
Aims: The aims of this study is to characterize the mechanism of vancomycin resistance in clinical Enterococcus faecium isolate recovered from ICU patient in Al-Nour Specialized Hospital in Makkah. Methods: In this study, resistant vancomycin resistant isolate was studied using whole genome sequencing. The genome was sequenced using Illumina MiSeq and sequence data was used to predict both the antibiotics resistant genes and the sequence type. In addition, the sequence data was used to screen for plasmids and virulence genes. Results: This resistant isolate were found to be belonging to sequence type 80 and harboring all known VanA genes, which include VanA, VanX, VanH, VanR, VanS, VanY and VanZ genes. Conclusion: Whole genome sequence of Vancomycin resistant E. faecium clinical strain revealed that it belonged to ST-80 which is a worldwide distributed sequence type. The vancomycin resistance was found to be due to the presence of VanA gene cluster. Three plasmids were predicted, two of them reported from E. faecium strains in many countries. Relation between transposon Tn-1546 and E. faecium plasmids also have been reported. Keywords: Enterococcus faecium, Vancomycin resistant, Whole genome sequencing | ||||||
|
INTRODUCTION
| ||||||
|
Antimicrobial resistance is a rapidly evolving problem associated with high cost and high mortality rates [1], [2]. Although, gastrointestinal tract of human consider to be the natural habitat of Enterococci, now it has become evident that environmental habitats are significant source of infection [3] . Like other opportunistic bacteria, enterococci causes infection when it finds a chance to reach other body sites. Enterococci can cause infections in urinary tract and cardiovascular system. In addition, enterococci can infect burns and wounds, abdomen, biliary tract, catheters, and other implanted medical devices [2]. Genus Enterococcus naturally resistant to many antibiotics such as cephalosporin, co-trimoxazole and aminoglycoside [4]. Several properties contributed to increase infection of multi-drug resistant enterococci including exposure to broad-spectrum antibiotics, ability to acquire new genetic characteristics and possibility to live in hospital settings. Increased antibiotic consumption plays a major role in acquiring antibiotic resistance to Enterococci [5]. Antibiotics used for infection prevention and growth promotion in food industry also play a role in resistance [6]. VRE are linked with high incidence of mortality in hospitals especially in very ill patients in intensive care units [7] and of resistant isolates recovered from intensive care units and surgical wards [8], [9]. Few studies have reported VRE from Saudi Arabia. In one study, the frequency of VRE in the gut microflora has been reported using conventional cultural methods and antimicrobial susceptibility testing. In a latter study, the VRE were identified in only six patients out of 4276 patients [10]. The second study characterized 34 vancomycin-resistant VanA E. faecium isolates obtained from two hospitals in Saudi Arabia, MLST of 33 isolates as determined by PFGE revealed that these isolates belonged to clonal complex CC17 [11]. | ||||||
|
MATERIALS AND METHODS
| ||||||
|
This study was carried out in Makkah Holy City, Saudi Arabia to study Enterococcus faecium clinical isolate from Al Nour Specialized Hospital.
DNA Extraction and Genome
Sequencing
The quality of the pair ends sequence reads were checked by FastQC before sequence assembly (BaseSpace Labs, Illumine Inc., USA). De novo assembly of (VRE) genomes was done using DNASTAR SeqManNGen 13.0.0 (DNASTAR, Madison, USA) using default settings, which included trimming of low quality sequences ends. Assembled genome was used for species identification and genome characterization. Species identification, Multi Locus Sequencing Typing, identification of plasmid, virulence factors and antibiotics resistant genes were predicted using Bacterial Analysis Pipeline from GoSeqIt (GoSeqIt, Denmark). | ||||||
RESULTS | ||||||
|
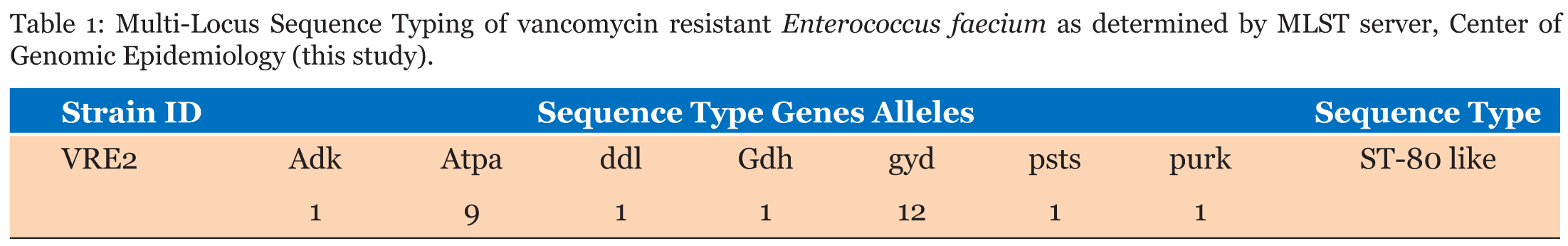
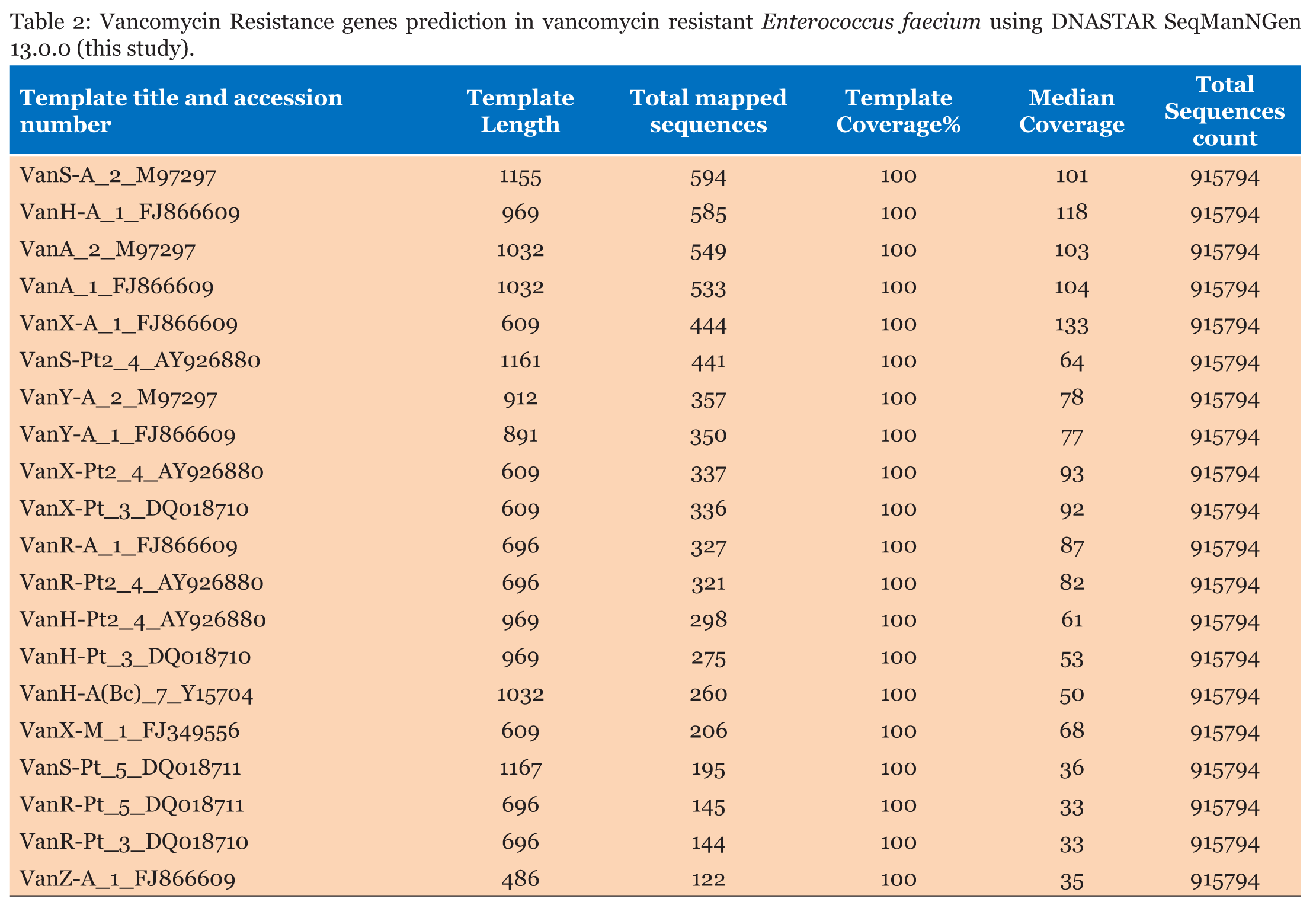
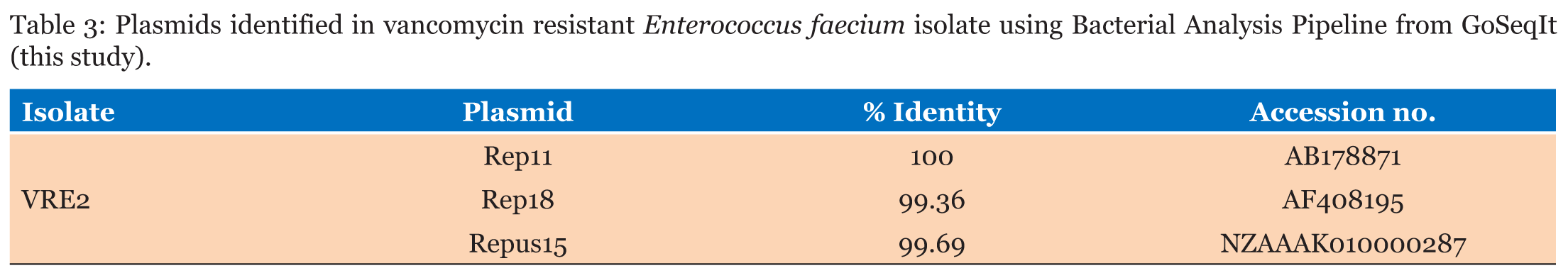
Isolated bacteria on MacConkey agar was identified based on cultural characteristics as Enterococci. Antimicrobial susceptibility testing showed that isolate was resistant to vancomycin. DNA was sequenced and sequencing reads were assembled (de novo assembly) and resulted in 600,797 assembled sequences forming 818 contigs. Average quality of assembled sequences was 35 and average coverage was 41X. Based on the 16SrRNA sequences, the isolate was found to be E. faecium determined by the Bacterial Analysis Pipeline from SeqIt . Multi-Locus Sequence Typing using Center of Genomic Epidemiology showed that E. faecium strain is belonging to ST-80 like (Table 1). Raw reads mapping using DNASTAR bioinformatics software revealed the presence of VanA, VanX, VanH, VanS, VanY and VanZ vancomycin resistance genes (Table 2). Three plasmids were identified using the Bacterial Analysis Pipeline from GoSeqIt (Table 3) | ||||||
| ||||||
| ||||||
| ||||||
|
DISCUSSION
| ||||||
|
Using whole genome sequencing, we are able to document the presence of vancomycin resistant E. faecium belonging to ST-80 in one of the major hospitals in Makkah, Saudi Arabia. Previous studies from Saudi Arabia reported emergence of VRE nosocomial infections from different hospitals. However, in other studies only PCR and PFGE were used to investigate vancomycin resistance mechanism and to trace the strains epidemically [11], [12], [13], [14], [15]. ST-80 is included in Clonal Complex 17 (CC17) lineage as found in clinical, environmental and animal samples from Tunisia, UK, Denmark, Canada, Portugal, Sweden and Korea [16], [17], [18], [19], [20], [21], [22]. Based on the epidemiological data provided by PubMLST, ST-80 isolated from blood sample of hospitalized patient in Israel in 1997 then appear in UK, South Korea (sample type is unknown). From 1999 until 2004 this sequence type appeared in Germany and Hungary (blood samples from hospitalized patients). In 2005, ST-80 was reported from skin and abdominal drainage of hospitalized patients in Italy, from blood samples of hospitalized patients in Sweden, Denmark and Canada. In 2014, this sequence type was reported from Russia (blood samples from hospitalized patients). In 2015 and 2016, this sequence type was reported in Algeria from infected surgical wounds and peritoneal pus of hospitalized patients. Most of the isolates belonging to this sequence type were vancomycin resistant [23]. Our study showed presence of VanA, VanX, VanH, VanR, VanS, VanY and VanZ genes which known to confer vancomycin resistance. Our findings are consistent with other studies that reported VRE infections caused by E.faecium are carrying VanA [24], [25], [26], [27], [28], [29], [30]. We are to document the presence of transposon Tn1546, which known to carry VanA, VanX, VanH, VanR, VanS, VanY and VanZ genes. Transposon Tn1546have different insertion sequences. Insertion element IS1216V is one of the most frequently detected insertion sequences within the transposon Tn1546, which was reported in VRE isolates from all over the world, in transposons containing vanA from different sources [28], [29], [30], [31]. Studies from Saudi Arabia also approved that transposon Tn1546 containing IS1216V has been associated with CC17 lineage [11],[15]. This insertion element has been detected at different positions and orientations in association with deletion of VanY and duplication of VanZ [32], [33], [34],[35]. our findings indicate presence of VanY and VanZ in existence of IS1216V, our findings are similar to Chao et al. [22]. Some studies found that deletion of VanY gene associated with presence of insertion sequence IS-1216V, however, our findings showed that VanY gene has been found together with the existence of IS-1216V [36]. Plasmids as carrier for VanA determinants played major role in spreading glycopeptides resistance [35]. Our results show that E. faecium contains plasmid replicons of different families, rep11 (repA-pB82) and rep18 (repA-pEF418 ) plasmids, recovered also from Portugal, Canada, Denmark, Germany, Italy, Spain, Sweden, Norway, Netherlands and Poland [32],[37], [38], [39],[40]. Plasmid rep11 (repA-pB82 ) is associated with human colonizing isolates . The high abundance of this plasmid family in E. faeciumis important to the development of these important clinical lineages of E. faecium [38]. Plasmid rep18 (repA-pEF418) reported on this study was found to be related to Tn1546 as mentioned above [32]. | ||||||
|
CONCLUSION
| ||||||
|
Whole genome sequencing of Vancomycin resistant E. faecium clinical strain revealed that it’s belonged to ST-80 which is a worldwide distributed sequence type. The vancomycin resistance was found to be due to the presence of VanA gene cluster. Three plasmids were predicted, two of them reported on E. faecium strains in many countries. Relation between transposon Tn-1546 and E. faecium plasmids also have been reported. | ||||||
|
REFERENCES
| ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
|
|
Author Contributions
Husam Aldeen Hassan Tahir – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Abdalla Ahmed – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Kawthar Abdelgaleil Mohammed Salih – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Bashir Sirag – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published |
|
Guarantor of Submission
The corresponding author is the guarantor of submission. |
|
Source of Support
None |
|
Consent Statement
Written informed consent was obtained from the patient for publication of this study. |
|
Conflict of Interest
Author declares no conflict of interest. |
|
Copyright
© 2018 Husam Aldeen Hassan Tahir et al. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information. |
|
|