|
|
Original Article
| ||||||
| Prevalence of methicillin-resistant Staphylococcus aureus isolates and their antibiotic susceptibility in tertiary care hospitals, India | ||||||
| Jeza Muhamad Abdul Aziz1, Francisca Kalavathi1 | ||||||
|
1Department of Microbiology, Bangalore City College, Bangalore University, Bangalore, India
| ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
[Print This Article]
[Similar article in Pumed] [Similar article in Google Scholar] |
| How to cite this article |
| Abdul Aziz JM, Kalavathi F. Prevalence of methicillin-resistant Staphylococcus aureus isolates and their antibiotic susceptibility in tertiary care hospitals, India. Edorium J Microbiol 2017;3:18–23. |
|
Abstract
|
|
Aims:
The broad range antibiotic resistance of Staphylococcus aureus (S. aureus) is a significant global threat to treatment of health care and community-associated infections. This study was aimed the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) and the antibiotic susceptibility pattern of S. aureus isolates in tertiary care hospitals in Bangalore, India from June 2012 to February 2013.
| |
|
Keywords:
Linezolid, Methicillin-resistant Staphylococcus aureus, Multidrug resistance, Prevalence
| |
|
Introduction
| ||||||
|
Staphylococcus aureus is a facultative anaerobic Gram-positive cocci. Clinically, it is the most frequently isolated human bacterial pathogen [1][2]. Staphylococcus aureus colonizes 30–50% of the human body in healthy individuals; sites include the nasal cavity, skin, gastrointestinal system, anus, and vagina vulva. Staphylococcus aureus also frequently causes a broad range of illnesses, ranging from minor skin infections like pimples, impetigo, and boils (furuncles) to life-threatening diseases including osteomyelitis, meningitis, pneumonia, endocarditis, toxic shock syndrome, sepsis, and bacteremia [2][3][4][5]. These infections are caused by hospital-associated methicillin-resistant S. aureus and community-associated MRSA. Hospital-acquired infections typically result from prolonged hospitalization, burns, trauma, chronic infection, lack of awareness, and contact with colonized patients [6] [7]. A survey in Taiwan in 1998 revealed that MRSA strains accounted for 84% of hospital-acquired S. aureus isolates and 45% of non-hospital acquired S. aureus isolates [8]. The prevalence of MRSA varies geographically within nations and between countries. A recent multicenter study from various regions in India reported the maximum prevalence of MRSA of 60–68% in tertiary care centres in central, south, and east India, and 54–57% in hospitals of west, south, and north India [9]. A study from Pakistan reported the highest prevalence of MRSA in Lahore (61%), followed by Karachi (57%), Rawalpindi, Islamabad (46%), Peshawar (36%), Azad Kashmir (32%), and Quetta (26%), with MRSA isolated from only 2% of sites in Sukkur [10]. The mecA gene, which is located on a mobile genetic element of the staphylococcal cassette chromosome (SCC), is responsible for the production of an abnormal penicillin binding protein designated PBP2a. PBP2a has decreased binding affinity for beta (ß)-lactam antibiotics, which results in resistance to methicillin and all ß-lactam antibiotics, including penicillin and cephalosporin [11]. The mecA gene complex contains insertion sites for plasmids and transposons, which promote the acquisition of resistance to other antibiotics. Therefore, cross-resistance is common to non-ß-lactam antibiotics, such as clindamycin, erythromycin, ciprofloxacin, co-trimoxazole, and gentamicin [12]. The treatment challenge posed by MRSA has become a serious therapeutic problem worldwide. These infections are a burden for patients and healthcare systems because of their associated high morbidity, mortality, and increased hospitalization costs [13]. Little is known about the prevalence of MRSA in Bangalore, India. The aim of the present study is to determine the prevalence of MRSA and the antibiotic susceptibility of S. aureus isolated from different clinical specimens in tertiary care hospitals in Bangalore, India. The goal is to identify the most appropriate antibiotic for treatment of S. aureus infections. | ||||||
|
Materials and Methods
| ||||||
|
Sample collection | ||||||
|
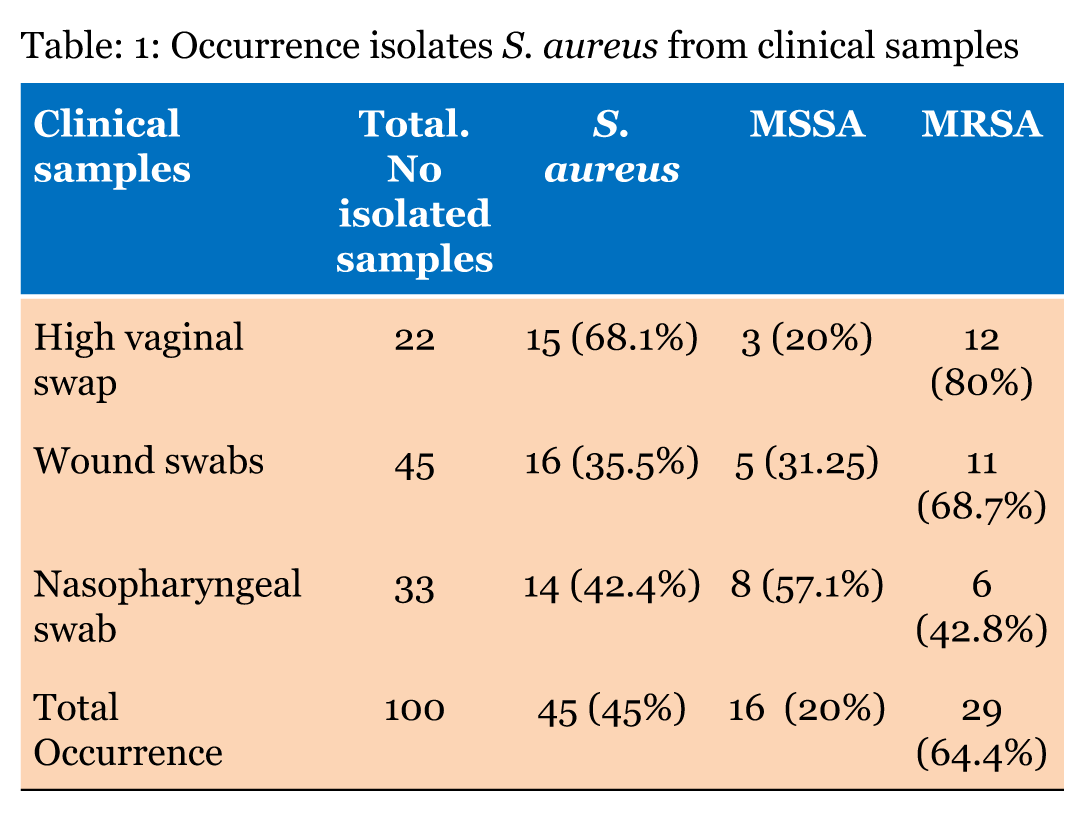
The standard technique [14] was used to collect 100 clinical swab samples, 22 high vaginal swabs, 45 wounds, and 33 nasopharyngeal sites (Table 1) from patients treated in tertiary care hospitals in Bangalore (India) from June 2012 to February 2013. Swab samples were taken after ethical approval and permission from each hospital. Sterile cotton-tipped swabs were used to collect the samples. Each swab was placed in an Amies transport medium tube and keep the medium cool by placed in a cool box when it transported to the lab according to the manufacturer’s instruction of (HiMedia Laboratories, India).
| ||||||
| Microbiological identification of isolated S. aureus | ||||||
|
Tryptic Soy broth (TSB; HiMedia Laboratories) was used for culture enrichment. Each cotton tip was broken off in a tube of TSB and the broth was vortexed for 10s to release bacteria from the swab into the broth. Each sample was incubated at 37°C for 48 h. A fresh sterile cotton swab was dipped into the culture and then used to inoculate a mannitol salt agar (MSA; HiMedia Laboratories) plate. In this procedure, the swab was pressed against the tube containing the TSB culture and rotated to remove excess fluid prior to streaking the entire surface of the MSA. The agar was then aerobically incubated for approximately 48 h at 37°C before examination for the visual indication of the fermentation of mannitol [15]. Colonies that developed were used to prepare Gram stains that were examined by light microscopy for the presence of Gram-positive cocci arranged in grape-like clusters, which is a hallmark of S. aureus. These Gram-positive colonies were inoculated into blood agar (BA; HiMedia Laboratories) and TSA plates, which were incubated aerobically at 35°C for 48 h. After incubation, the developed colonies were examined for morphological characteristics e.g., smooth, convex, circular, yellow colonies) and the presence of ß-haemolysis. Moreover, to confirm S. aureus, the coagulase and catalase tests were performed using samples from BA and TSA, respectively. The BA colonies were not used for the catalase test, because red blood cells contain catalase, leading to a false positive reaction [15][16]. | ||||||
| Phenotypic and genotypic identification of MRSA | ||||||
|
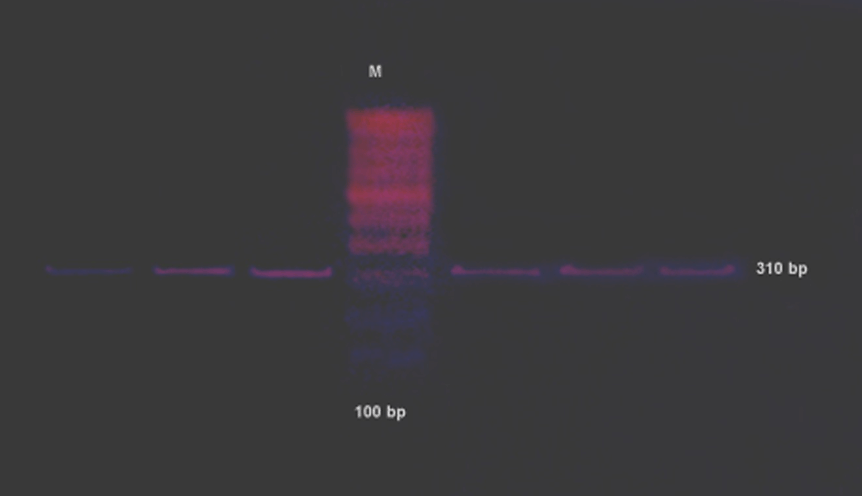
Sterile swabs were used to obtain colonies of confirmed S. aureus from BA. Each sample was suspended in a single vial filled with 5 ml of TSB. The turbidity was adjusted to a McFarland standard of 0.5, which corresponds to 1x108 colony forming units (CFU)/ml). Then, another sterile cotton swab was dipped into the suspension and swabbed on the surface of Mueller-Hinton agar (MHA; HiMedia Laboratories). The plates were kept at room temperature for 15 min [17]. Confirmed S. aureus strains (n = 45, 45%) were phenotypically screened for mecA by the addition of a disk containing cefoxitin 30 µg (HiMedia Laboratories) to the surface of the inoculated MHA, followed by incubation at 30°C for 24 h [18]. After the incubation period, the diameters of the inhibition zones were measured. The results were recorded based on interpretive criteria according to the recommendations in the Clinical and Laboratory Standards Institute (CLSI) guidelines [17]. Confirmation was provided by PCR-amplification of mecA. Bacterial genomic DNA was extracted using cetyltrimethylammonium bromide [19]. The DNA was suspended in 200 µl TE buffer, ph 7.4 (Promega Corporation), and stored at -20°C until needed for analyses. The DNA concentration in suspension was measured using a nanodrop-100 spectrophotometer (Thermo Fisher Scientific, USA) [20]. The mecA forward primer (5'-GTA GAA ATG ACT GAA CGT CCG ATA A) and reverse primer (5'-CCA ATT CCA CAT TGT TTC GGT CTAA) were used to amplify 310 bp of mecA MRSA by PCR [21]. The 25-µl PCR reaction mixture contained 5 µl DNA template, 2 µl of each primer (20 µM), 2.5 µl of 10x buffer, 2.5 µl MgCl2, 1 µl of Taq DNA polymerase (Bangalore Genei), 4 µl of deoxynucleoside triphosphates (DNTPS), and 6 µl distilled water. A thermal cycler (Corbett Life Science, USA) was programmed with the initial denaturation,4 min at 94°C; 30 cycles with a 45-s denaturation step at 94°C, a 45-s annealing step at 56°C and a 30-s extension step at 72°C and 2 min extension step at 72°C and a holding step at 4°C to amplify DNA in an Eppendorf tube ( HiMedia Laboratories) [22]. Eight microliters of each PCR product was loaded on to a 1.0% agarose gel with 2 µl ethidium bromide. Resolved bands were visualized using an ultraviolet illuminator. | ||||||
| Antibiotic susceptibility testing | ||||||
|
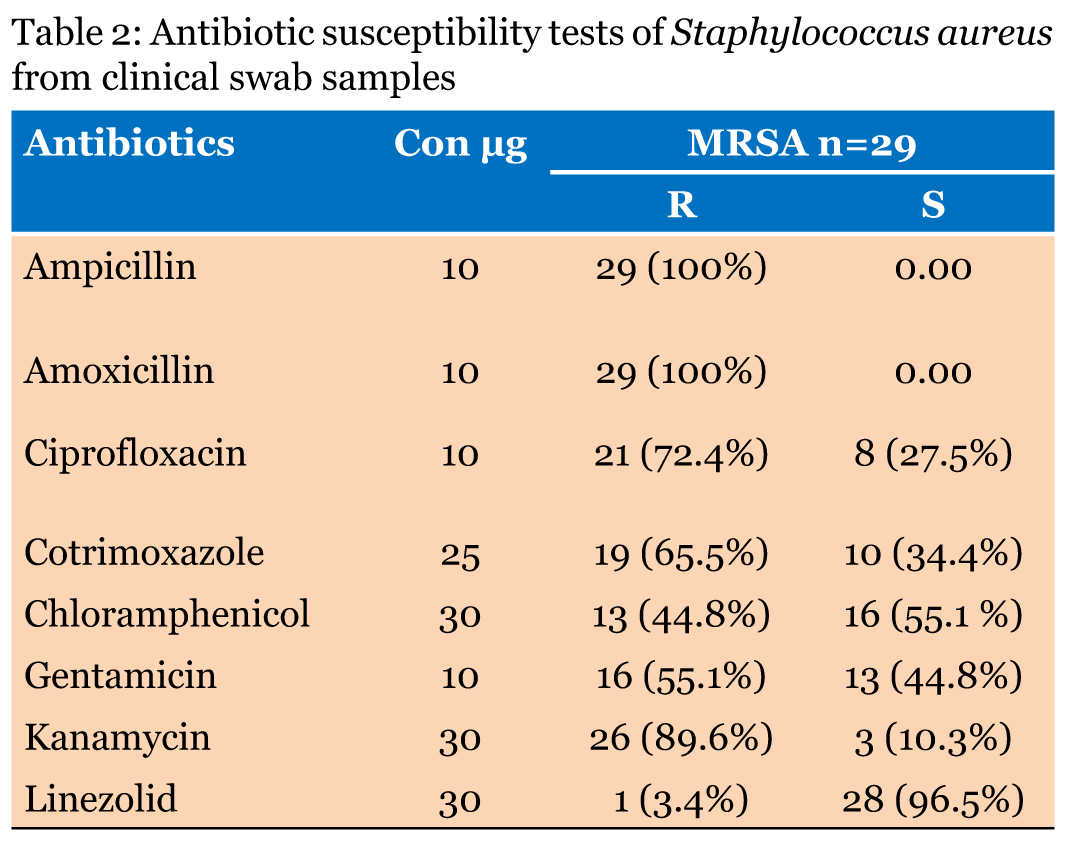
Susceptibility testing of MRSA isolates for eight antibiotics was performed using the Kirby-Bauer disk diffusion method on MHA as recommended in the CLSI protocol [17]. All antibiotic disks were obtained from HiMedia Laboratories. These included inhibitors of cell wall synthesis [ampicillin (10 µg/disk) and amoxicillin (10 µg/disk)]; inhibitors of protein synthesis [linezolid (30 µg/disk), gentamicin (10 µg/disk), and kanamycin (30 µg/disk)]; and inhibitors of DNA synthesis [ciprofloxacin (10 µg/disk), chloramphenicol (30 µg/disk), and cotrimoxazole (25 µg/disk]. Antibiotic disks were placed on MHA by using sterile forceps and were gently pressed down to ensure contact. Within 15 min of the disks being placed, the plates were inverted and incubated aerobically at 37°C for 18 h. Staphylococcus aureus ATCC 43300 was used as the quality control strain for disk diffusion. After incubation, the diameter of the inhibition zone was measured, and the results were recorded based on interpretive criteria according to the recommendations in CLSI guidelines [17]. | ||||||
| ||||||
|
Results | ||||||
|
Of the 100 isolates, 80 (80%) were identified to be Gram-positive cocci. Of these, 72 showed positive results in the catalase test for Staphylococcus and Micrococcus spp. Of these 73 samples, 45 were identified as S. aureus based on the fermentation of mannitol, which was evident on MSA as a colour change from red to yellow, and on the positive result in the coagulase test. Of the 45 isolates, the cefoxitin disk diffusion test identified 16 as being sensitive to methicillin (i.e. MSSA) and 29 as MRSA. To confirm the results of the biochemical tests, all S. aureus samples were analysed by polymerase chain reaction detection of mecA mediated resistance (Figure 1). Thus, both analyses revealed 29 (64.4%) MRSA from the S. aureus isolates. The 29 isolates were acquired from 12 high vaginal swabs, 11 wound swabs, and 6 nasopharyngeal swab specimens (Table 1). The antibiotic resistance patterns of the 29 MRSA isolates are summarized in (Table 2) A high level of resistance (100%) was evident for the penicillin group (amoxicillin and ampicillin), the fluoroquinolones group (ciprofloxacin, 72.4%), and the sulfonamide and trimethoprim group (cotrimoxazole, 65.5%). A low level of resistance was evident for the phenicol group (chloramphenicol, 44.8%). Moderate resistance to gentamicin (55.1%) and high resistance to kanamycin (89.6%) of the aminoglycoside group were observed, and resistance to linezolid was low (one isolate, 3.4%). The MRSA isolates tended to display a high level of resistance to diverse antibiotics, qualifying them as multi-drug resistant (MDR) MRSA. | ||||||
| ||||||
| ||||||
|
Discussion
| ||||||
|
In this study, 68.1% of the isolated S. aureus were from the high vagina region. Nkwenlag et al. [23] reported that 50.9% of S. aureus isolates in their study were from the genital area of healthy persons in hospitals in Cameroon. The prior and present findings may reflect the increased susceptibility of the female anatomy to bacterial colonization and infection, which could be attributed to poor personal hygiene or poor health education [24]. The phenotype-based detection of MRSA in the present study involved the cefoxitin disk diffusion method. The method identifies MRSA with high sensitivity because of the relatively better detection of PBP2a production in S. aureus strains harbouring mecA than the previously used oxacillin disk diffusion method [18][25][26]. The molecular (PCR) detection results of the MRSA mecA gene of MRSA corroborated the phenotypic analysis results; however, PCR is the preferred approach because of its efficiency [27][28]. The prospective analysis of 45 S. aureus isolates revealed the highest prevalence (64.4%) of MRSA within eight months in tertiary care hospitals in Bangalore, India which is higher compared to the previous study conducted in other regions of India, [29] and lower than the rate of 38.44% reported from a hospital in Delhi, India [30] and the rates of 31.1% from multiple sites in Tamil Nadu [31] and 80.89% in Indore [32]. The collective results confirm that the prevalence of MRSA varies between different parts of the country and between tertiary care centres in the same geographical region. For a long-time, penicillin has been the main stay for the management of a variety of staphylococcal infections. However, staphylococci have gradually acquired penicillin resistance. This is clearly exemplified in the current study, where 100% resistance was evident against ampicillin and amoxicillin. Hanumanthappa et al. [33] reported the same findings in 2003 from Karnataka, India. The higher prevalence of antibiotic resistance in India may reflect the misuse or abuse of penicillin antibiotics. In the present study, 89.6% of S. aureus strains were resistant to kanamycin, which is lower than the result reported by Shittu et al. [34]. The resistance of S. aureus isolates to ß-lactam antibiotics is related to the production of ß-lactamase. The enzyme hydrolyses the ß-lactam ring of the relevant antibiotics, destroying their antimicrobial activity [2]. Presently, the S. aureus isolates displayed resistance rates of 55.1% and 65.5% to gentamicin and cotrimoxazole, respectively. However, in a tertiary referral hospital in eastern Uttar Pradesh, India, the resistance rate to these antibiotics exceeded 80%, [35][36] while Rajaduraipandi et al. [31] reported resistance rates of 62% and 63.2% to gentamicin and cotrimoxazole, respectively, in southern districts of Tamil Nadu, India. Previous studies reported MRSA isolates in India that harboured SCCmec type IV or V resistance to gentamicin [37][38]. Presently, 55.1% of MRSA isolates were resistant to gentamicin. The collective findings indicate the community dispersion of MRSA SCCmec type IV strains in Bangalore, India. Presently, 72.4% of the MRSA isolates displayed resistance to ciprofloxacin. Previous studies reported lower rates (40–48%) of resistance to ciprofloxacin in a tertiary care hospital in India [29][39]. The present chloramphenicol resistance rate of 60% is markedly different from the 38.1% reported from Nigeria by Onwubiko et al. [40]. Drug resistance could be associated with earlier exposure of these drugs to isolates, which may have enhanced the development of resistance. The most effective chemotherapeutic agent observed against MRSA in this study was linezolid. The 97.61% sensitivity observed is similar to that reported by Rajaduraipandi et al. [31] in 2006. However, it is somewhat lower than that reported by Hasani et al. [41] in 2013 in Iran. The MDR character of the MRSA isolates (Table 2) could be a result of the use of inappropriate antimicrobials because of the widespread availability of antimicrobials without a prescription or those prescribed by non-skilled practitioners. The consequences of using these antimicrobials can be inadequate therapy and further drug resistance. | ||||||
|
Conclusion
| ||||||
|
Methicillin-resistant Staphylococcus aureus comprised 64% of the S. aureus isolates, which represents a high prevalence, and treatment of infections caused by S. aureus with ß-lactam antibiotics and other drug groups can be undependable. The cefoxitin disk diffusion test was revealed to be an accurate phenotype-based method for the detection MRSA in a laboratory where polymerase chain reaction is not available. The most significant principal finding was the multidrug resistance nature of the S. aureus isolates, except for linezolid, which can be considered as the drug of choice for treatment of MRSA infections. Furthermore, the incidence of MRSA infection can be reduced by monitoring the antibiotic susceptibility of MRSA isolates, identification of MRSA infections within the hospital, and increasing awareness of MRSA in the general public. Further studies will be required to confirm the present results. | ||||||
|
References
| ||||||
| ||||||
|
[HTML Abstract]
[PDF Full Text]
|
| Acknowledgements |
|
We gratefully acknowledge Bangalore City College for providing laboratory facilities for this work. |
|
Author Contributions
Jeza Muhamad Abdul Aziz – Substantial contributions to conception and design, Acquisition of data, Analysis and interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published Francisca Kalavathi – Substantial contributions to conception and design, Analysis and interpretation of data, Revising it critically for important intellectual content, Final approval of the version to be published |
|
Guarantor of Submission
The corresponding author is the guarantor of submission. |
|
Source of Support
None |
|
Conflict of Interest
Authors declare no conflict of interest. |
|
Copyright
© 2017 Jeza Muhamad Abdul Aziz et al. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information. |
|
|